Synthetic Tests
[1]:
import numpy as np
import pandas as pd
import requests
import io
from scipy import interpolate
import matplotlib.pyplot as plt
from cartopy import crs as ccrs, feature as cfeature
import json
import ast
from gen_synth_data import gen_data, distort_data
from sedprep.kalman_filter import Filter
from sedprep.utils import lif
from sedprep.data_handling import add_identifier, read_arch_data, clean_data
from sedprep.constants import field_params
from sedprep.plot_functions import plot_DIF
folder = "../dat"
cpu
[2]:
np.random.seed(1000)
new_sample_from_prior = False # set to True to generate a new reference model
# We use the temporal and spartial distribution of real archaeo data and the sed_data_KLK.csv
pre = "https://nextcloud.gfz-potsdam.de/s/"
rejection_response = requests.get(f"{pre}WLxDTddq663zFLP/download")
rejection_response.raise_for_status()
with np.load(io.BytesIO(rejection_response.content), allow_pickle=True) as fh:
to_reject = fh['to_reject']
data_response = requests.get(f"{pre}r6YxrrABRJjideS/download")
arch_real = read_arch_data(io.BytesIO(data_response.content), to_reject)
# arch_real = pd.read_csv("../dat/synth_data/arch.csv")
sed_real = pd.read_csv("../dat/P3.csv")
knots, dt = np.linspace(
min(arch_real["t"]), max(arch_real["t"]), 2000, retstep=True
)
knots = np.concatenate((
[min(knots) - 2], [min(knots) - 1], knots, [max(knots) + 1], [max(knots) + 2]
))
if new_sample_from_prior:
model = Filter(**field_params)
prior_sample = model.sample_prior(ts=knots, n_samps=10)[0, :, :]
np.save("../dat/prior_sample.npy", prior_sample)
else:
prior_sample = np.load("../dat/prior_sample.npy")
vals = np.insert(prior_sample.T, [prior_sample.T.shape[0], 0], [prior_sample.T[-1], prior_sample.T[0]], axis=0)
spline = interpolate.BSpline(knots, vals, 1)
Rejected 347 outliers.
/opt/conda/envs/virtualenv/lib/python3.12/site-packages/sedprep/data_handling.py:553: UserWarning: Records with indices [ 72 73 74 75 76 77 78 79 80 81 108 109
357 358 1065 1066 1153 2402 2575 2671 2753 2756 2757 3036
3215 3217 3218 3219 3220 3221 3222 3223 3224 3225 3503 3772
3988 4047 4235 4415 4504 4530 4531 4655 5172 5326 5434 5496
5551 5707 5763 5802 5805 5959 6053 6105 6647 6706 6781 6809
7149 7580 7585 7593 7692 7702 7735 7776 7806 7826 7881 7979
8067 8296 8327 8427 8464 8510 8670 8708 8752 9224 9653 10002
10132 10331 10509 10604 10647 11527 11758] contain declination, but not inclination! The errors need special treatment!
To be able to use the provided data, these records have been dropped from the output.
[3]:
np.random.seed(1000)
gen_new_arch_data = False # set to True to generate new synthetic arch data
if gen_new_arch_data:
arch, _ = gen_data(
t=arch_real["t"], # use temporal distribution of real arch data
lat=arch_real["lat"], # use spatial distribution of real arch data
lon=arch_real["lon"], # use spatial distribution of real arch data
field_spline=spline, # use previously generated prior sample
dt=arch_real["dt"], # use temporal errors of real arch data
ddec=arch_real["dD"], # use measurement errors of real arch data
dinc=arch_real["dI"], # use measurement errors of real arch data
dint=arch_real["dF"], # use measurement errors of real arch data
depth=None, # arch data does not has depth
with_noise=True, # add some noise to synthetic arch data
)
arch["type"] = "arch_data"
# # To use your new data store it as a csv file using
arch.to_csv(f"{folder}/arch.csv", index=False)
# # Then run the following line to add unique identifiers
add_identifier(f"{folder}/arch.csv")
else:
arch = pd.read_csv(f"{folder}/arch.csv")
[4]:
np.random.seed(1000)
gen_new_ref_sed_data = False # set to True to generate new reference sediment data
ref_sed_dfs = {}
ref_sed_NEZs = {}
lats = {"sweden": sed_real["lat"][0], "rapa": -27.616}
lons = {"sweden": sed_real["lon"][0], "rapa": -144.283}
for loc in ["sweden", "rapa"]:
if gen_new_ref_sed_data:
ref_sed, ref_sed_NEZ = gen_data(
t=sed_real["t"], # use temporal distribution of real sed data
lat=lats[loc], # use latitude distribution of real sed data (-27.616 for rapa)
lon=lons[loc], # use longitude distribution of real sed data (-144.283 for rapa)
field_spline=spline, # use previously generated prior sample
dt=sed_real["dt"], # use temporal errors of real sed data
depth=sed_real["depth"],# use depth of real sed data
with_noise=False, # no noise for reference process
)
# Store the data as a csv file using
ref_sed.to_csv(f"{folder}/ref_sed_{loc}.csv", index=False)
np.save(f"{folder}/ref_sed_{loc}_NEZ.npy", ref_sed_NEZ)
ref_sed_NEZs[loc] = np.load(f"{folder}/ref_sed_{loc}_NEZ.npy")
ref_sed_dfs[loc] = pd.read_csv(f"{folder}/ref_sed_{loc}.csv")
else:
ref_sed_NEZs[loc] = np.load(f"{folder}/ref_sed_{loc}_NEZ.npy")
ref_sed_dfs[loc] = pd.read_csv(f"{folder}/ref_sed_{loc}.csv")
[5]:
np.random.seed(1000)
gen_new_sed_data = False # set to True to generate new synthetic sed data
# since sed_real has no intensity errors we use the following
dint = np.random.normal(4, 2, len(sed_real)).clip(0.1, None)
# add two subsections
subsections={"A1": np.arange(0, 60, 1), "A2": np.arange(60, len(ref_sed_dfs["sweden"]), 1)}
bs_DIs = {"triangle": [10, 10, 0, 10], "square": [5, 1, 20, 1], "ramp": [1, 1, 0, 20]}
bs_Fs = bs_DIs
# offset parameters for two sections
offsets = {"A1": 30., "A2": -10.}
f_shallow=0.6
cal_fac = 90
sed_dfs = {}
for loc in ["sweden", "rapa"]:
ref_sed = ref_sed_dfs[loc]
ref_sed_NEZ = ref_sed_NEZs[loc]
for file in ["triangle", "square", "ramp"]:
name = f"{loc}_{file}"
if gen_new_sed_data:
sed = distort_data(
ref_sed_dfs[loc],
ref_sed_NEZs[loc],
ddec=sed_real["dD"], # use measurement errors of real sed data
dinc=sed_real["dI"], # use measurement errors of real sed data
dint=dint, # measurement errors for intensity
bs_DI=bs_DIs[file], # lock-in function parameters for directions
bs_F=bs_Fs[file], # lock-in function parameters for intensity
offsets=offsets, # declination offsets per sections
f_shallow=f_shallow, # inclination shallowing factor
cal_fac=cal_fac, # calibration factor for intensity
subsections=subsections,# subsections
)
sed["type"] = "sediments"
sed.to_csv(f"{folder}/sed_{name}.csv", index=False)
# Add unique identifiers
add_identifier(f"{folder}/sed_{name}.csv")
sed_dfs[name] = pd.read_csv(f"{folder}/sed_{name}.csv")
else:
sed_dfs[name] = pd.read_csv(f"{folder}/sed_{name}.csv")
[6]:
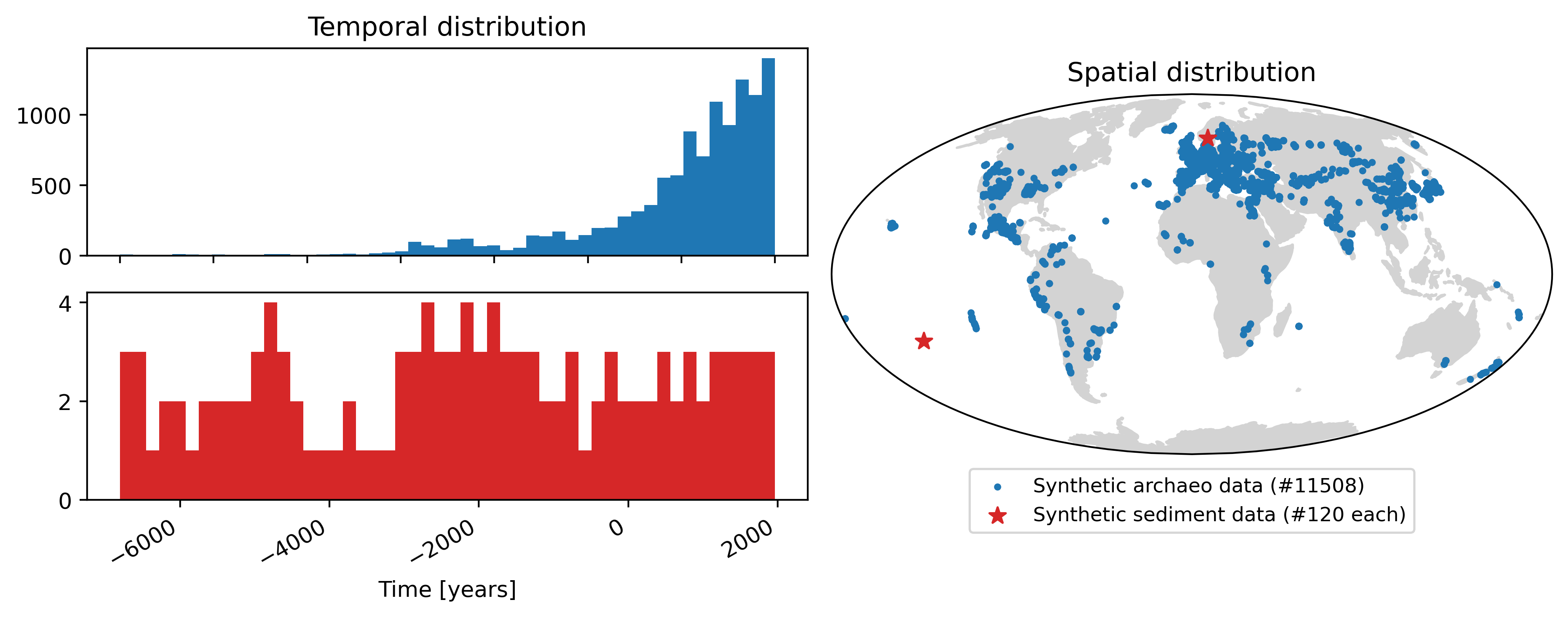
proj = ccrs.Mollweide()
fig = plt.figure(figsize=(10, 4), dpi=400)
arch_temp_dist = fig.add_subplot(2, 2, 1)
sed_temp_dist = fig.add_subplot(2, 2, 3)
sph_ax = fig.add_subplot(1, 2, 2, projection=proj)
arch_temp_dist.hist(arch.t, bins=50, color="C0")
arch_temp_dist.set_title("Temporal distribution")
sed_temp_dist.hist(ref_sed_dfs["rapa"].t, bins=50, color="C3")
sed_temp_dist.set_xlabel("Time [years]")
sph_ax.set_title("Spatial distribution")
sph_ax.set_global()
sph_ax.add_feature(cfeature.LAND, zorder=0, color="lightgray")
sph_ax.scatter(
arch.lon,
arch.lat,
c="C0",
label="Synthetic archaeo data (#" + str(int(len(arch))) + ")",
s=5,
alpha=1,
zorder=1,
rasterized=True,
transform=ccrs.PlateCarree(),
)
for i, sed_data in enumerate(
[ref_sed_dfs["sweden"], ref_sed_dfs["rapa"]]
):
sph_ax.scatter(
sed_data.lon[0],
sed_data.lat[0],
c="C3",
label="Synthetic sediment data (#" + str(int(len(sed_data))) + " each)" if i == 0 else None,
s=60,
marker="*",
alpha=1,
zorder=1,
rasterized=True,
transform=ccrs.PlateCarree(),
)
sph_ax.legend(loc="center", prop={"size": 9}, bbox_to_anchor=(0.5, -0.13), ncol=1)
fig.autofmt_xdate()
fig.tight_layout()
/opt/conda/envs/virtualenv/lib/python3.12/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/110m_physical/ne_110m_land.zip
[7]:
preprocess_data = False # set to True to apply estimated parameters to clean data
prep_dfs = {}
for loc in ["sweden", "rapa"]:
for file in ["triangle", "square", "ramp"]:
name = f"{loc}_{file}"
sed = pd.read_csv(f"{folder}/sed_{name}.csv")
res_DI = pd.read_csv(f"../results/sed_{name}_DI.csv")
res_F = pd.read_csv(f"../results/sed_{name}_F.csv")
est_bs_DI_4p = ast.literal_eval(res_DI[res_DI.optimizer=="scipy"].bs_DI.values[0])
est_bs_F_4p = ast.literal_eval(res_F[res_F.optimizer=="scipy"].bs_F.values[0])
est_bs_DI_2p = ast.literal_eval(res_DI[res_DI.optimizer=="scipy"].bs_DI.values[1])
est_bs_F_2p = ast.literal_eval(res_F[res_F.optimizer=="scipy"].bs_F.values[1])
est_offsets_4p = json.loads(res_DI[res_DI.optimizer=="scipy"].offsets.values[0].replace("'", '"'))
est_offsets_2p = json.loads(res_DI[res_DI.optimizer=="scipy"].offsets.values[1].replace("'", '"'))
est_f_shallow_4p = res_DI[res_DI.optimizer=="scipy"].f_shallow.values[0]
est_f_shallow_2p = res_DI[res_DI.optimizer=="scipy"].f_shallow.values[1]
est_cal_fac_4p = res_F[res_F.optimizer=="scipy"].cal_fac.values[0]
est_cal_fac_2p = res_F[res_F.optimizer=="scipy"].cal_fac.values[1]
if preprocess_data:
prep_dfs[name + "_4p"] = clean_data(
sed,
est_bs_DI_4p,
est_bs_F_4p,
est_offsets_4p,
est_f_shallow_4p,
est_cal_fac_4p,
)
prep_dfs[name + "_4p"].to_csv(
f"../results/preprocessed/sed_{name}_preprocessed_4p.csv", index=False
)
prep_dfs[name + "_2p"] = clean_data(
sed,
est_bs_DI_2p,
est_bs_F_2p,
est_offsets_2p,
est_f_shallow_2p,
est_cal_fac_2p,
)
prep_dfs[name + "_2p"].to_csv(
f"../results/preprocessed/sed_{name}_preprocessed_2p.csv", index=False
)
else:
prep_dfs[name + "_4p"] = pd.read_csv(f"../results/preprocessed/sed_{name}_preprocessed_4p.csv")
prep_dfs[name + "_2p"] = pd.read_csv(f"../results/preprocessed/sed_{name}_preprocessed_2p.csv")
[8]:
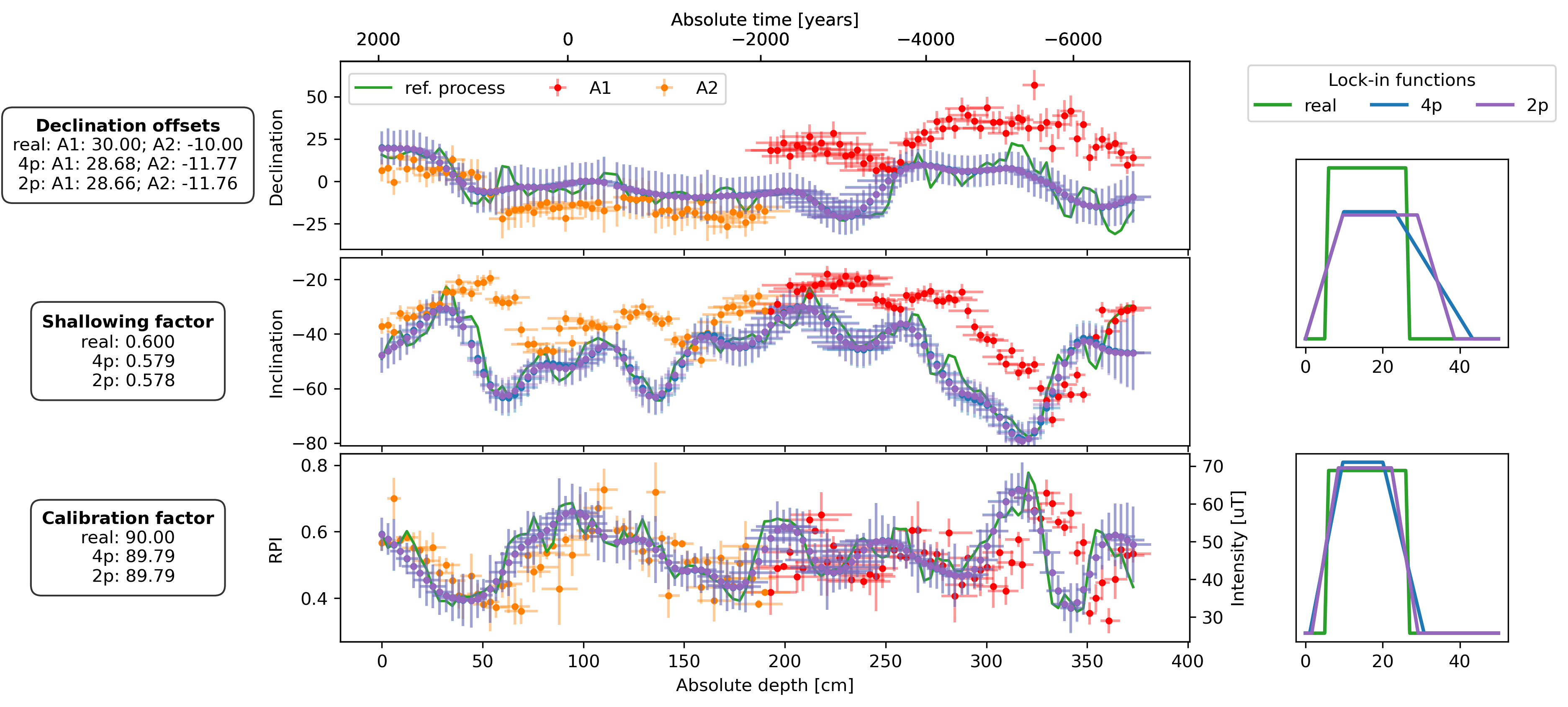
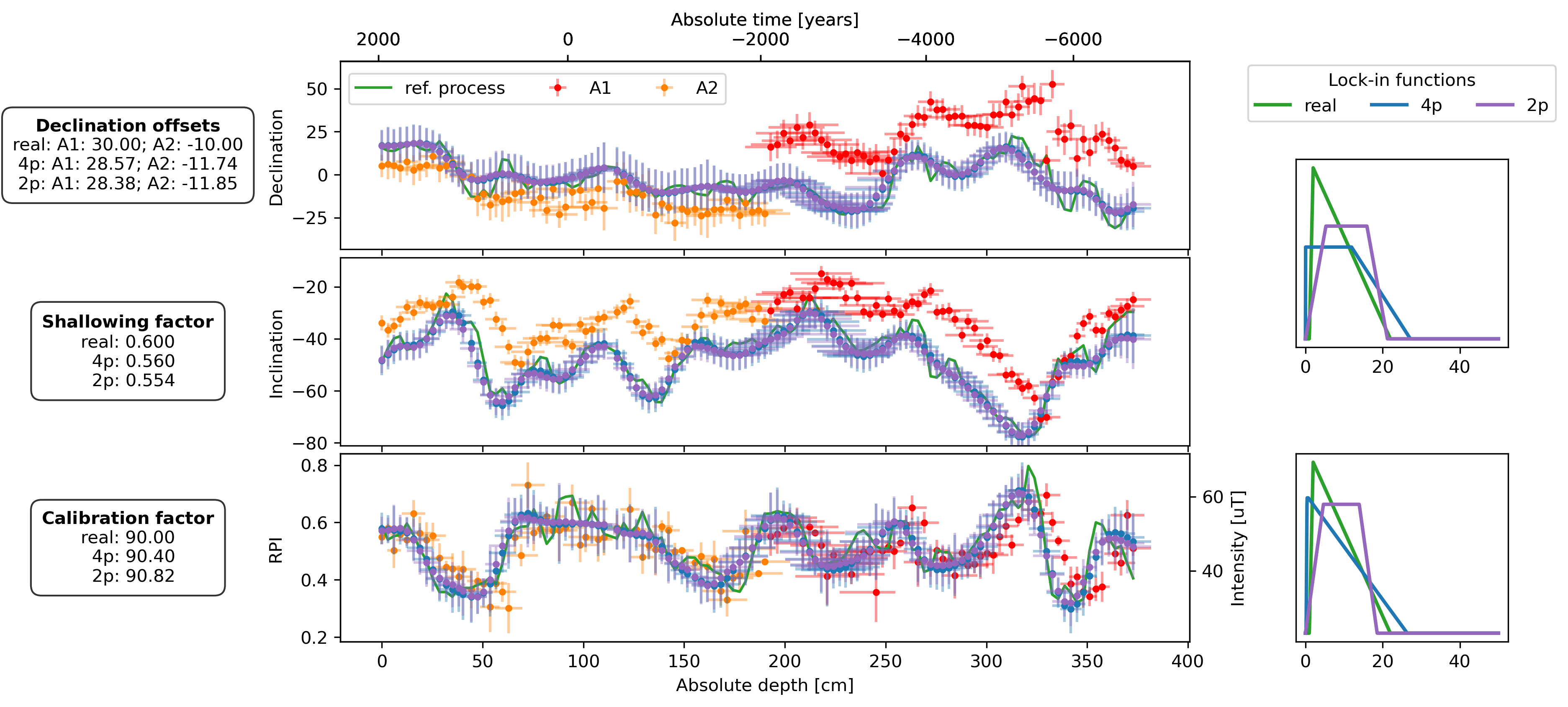
lif_knots = np.linspace(0, 50, 1000)
props = dict(boxstyle='round,pad=0.6', facecolor='none', alpha=0.8)
for loc in ["sweden", "rapa"]:
ref_sed = pd.read_csv(f"{folder}/ref_sed_{loc}.csv")
for file in ["triangle", "square", "ramp"]:
name = f"{loc}_{file}"
sed = pd.read_csv(f"{folder}/sed_{name}.csv")
res_DI = pd.read_csv(f"../results/sed_{name}_DI.csv")
res_F = pd.read_csv(f"../results/sed_{name}_F.csv")
est_bs_DI_4p = ast.literal_eval(res_DI[res_DI.optimizer=="scipy"].bs_DI.values[0])
est_bs_F_4p = ast.literal_eval(res_F[res_F.optimizer=="scipy"].bs_F.values[0])
est_bs_DI_2p = ast.literal_eval(res_DI[res_DI.optimizer=="scipy"].bs_DI.values[1])
est_bs_F_2p = ast.literal_eval(res_F[res_F.optimizer=="scipy"].bs_F.values[1])
est_offsets_4p = json.loads(res_DI[res_DI.optimizer=="scipy"].offsets.values[0].replace("'", '"'))
est_offsets_2p = json.loads(res_DI[res_DI.optimizer=="scipy"].offsets.values[1].replace("'", '"'))
est_f_shallow_4p = res_DI[res_DI.optimizer=="scipy"].f_shallow.values[0]
est_f_shallow_2p = res_DI[res_DI.optimizer=="scipy"].f_shallow.values[1]
est_cal_fac_4p = res_F[res_F.optimizer=="scipy"].cal_fac.values[0]
est_cal_fac_2p = res_F[res_F.optimizer=="scipy"].cal_fac.values[1]
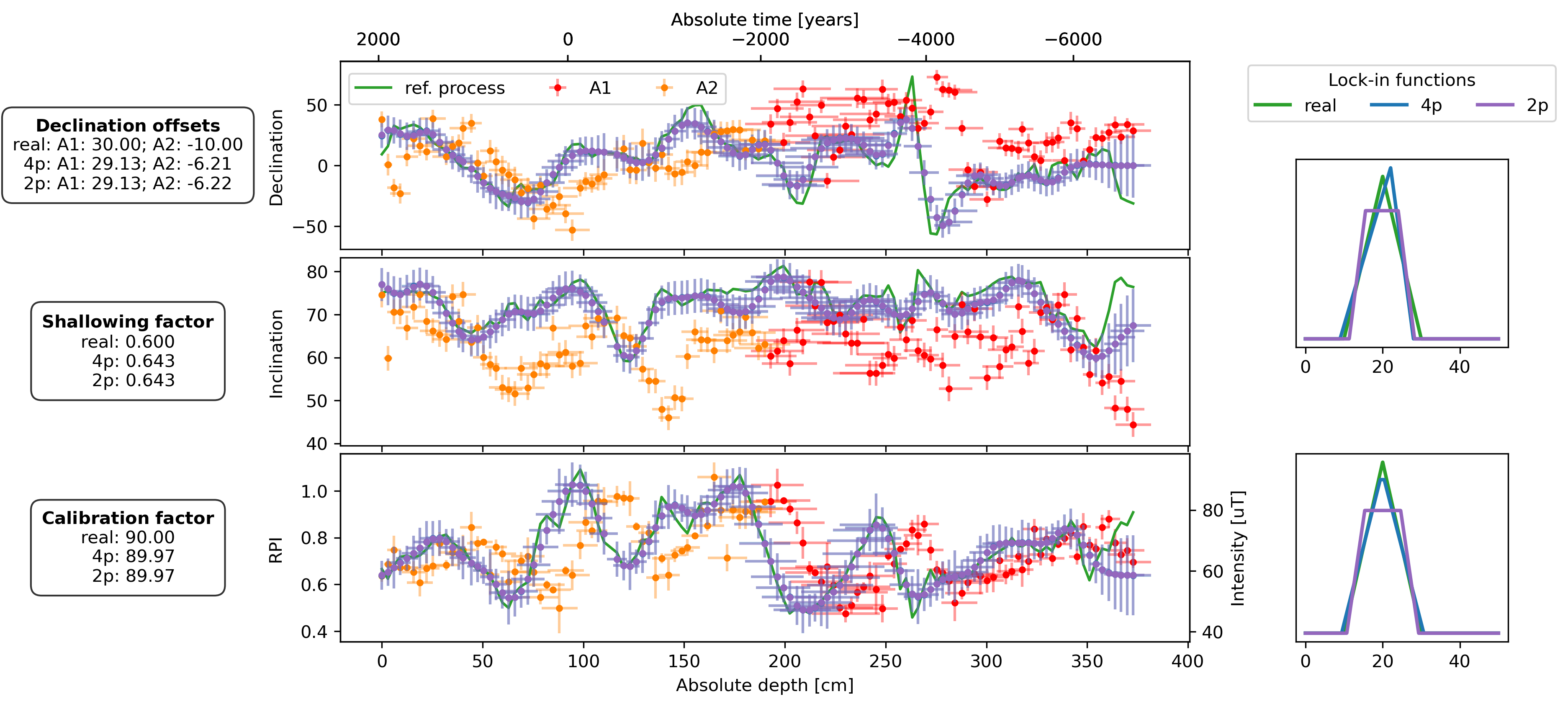
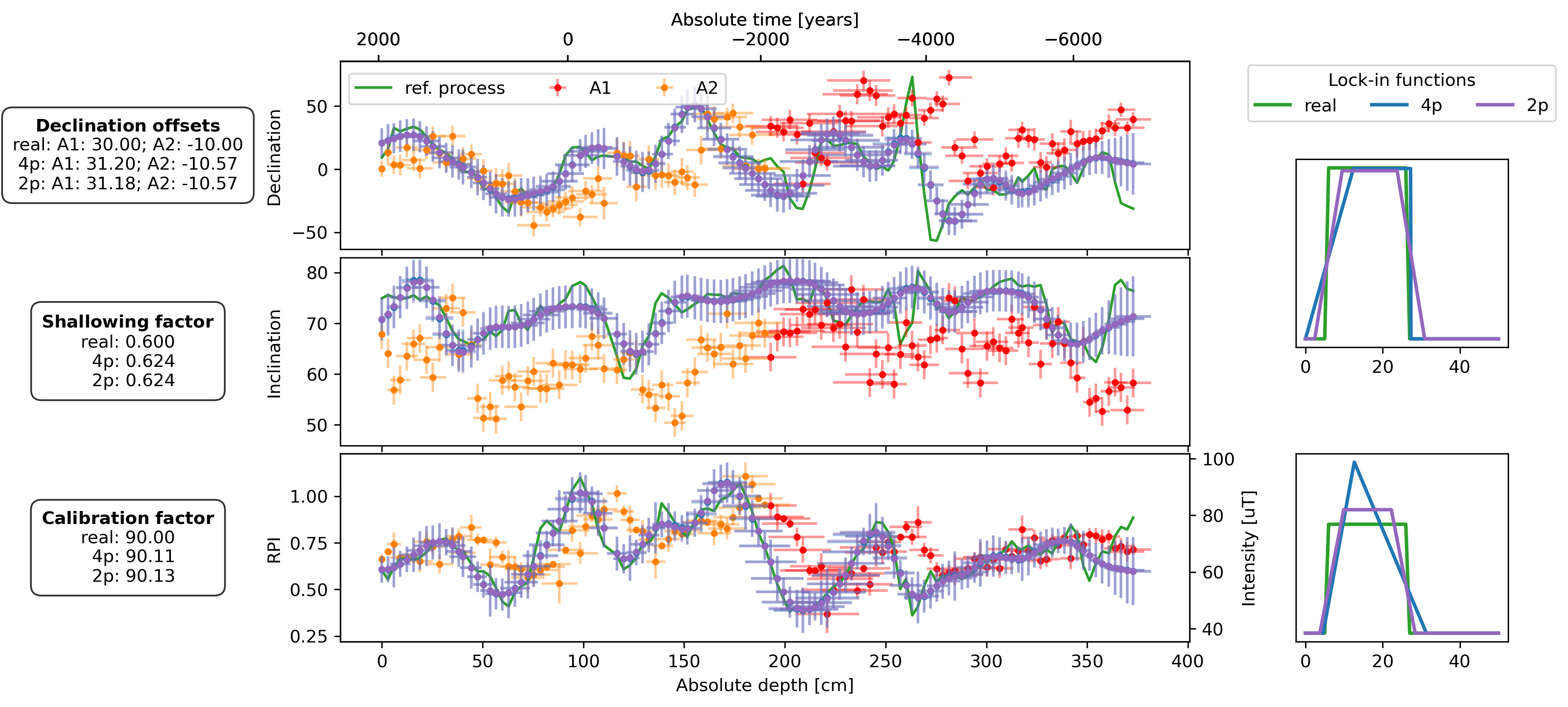
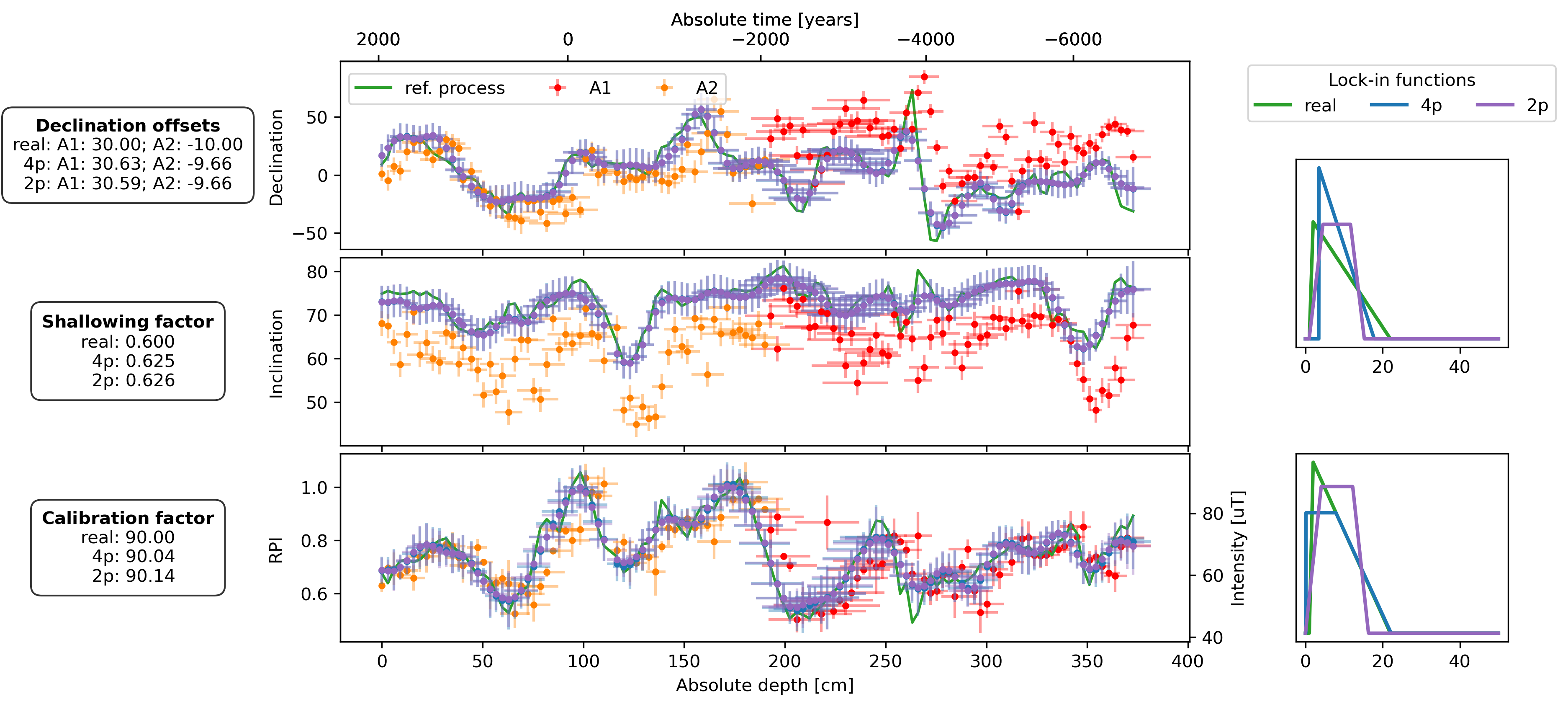
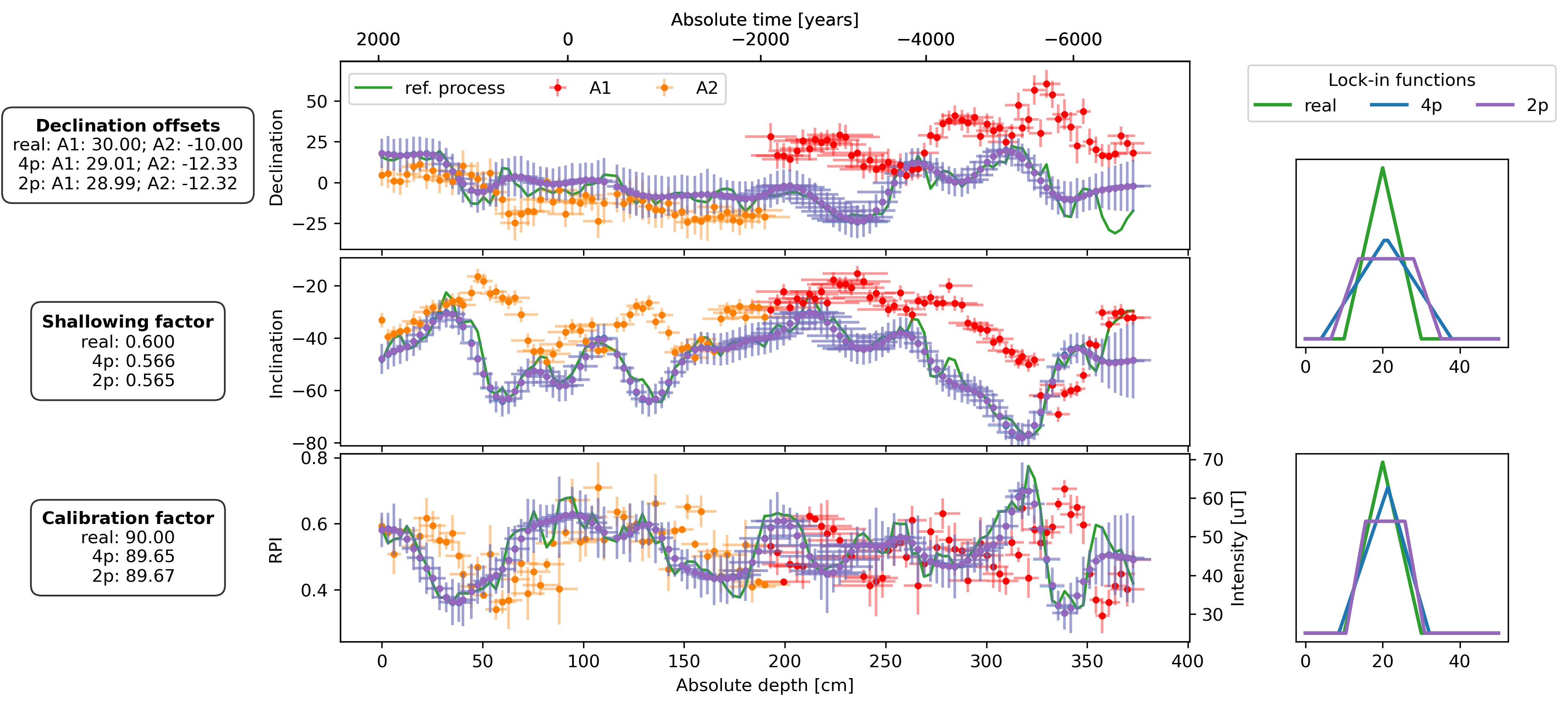
fig, axs = plt.subplot_mosaic(
2 * [3 * ["D"] + ["."]]
+ 2 * [3 * ["D"] + ["LIF_DI"]]
+ 2 * [3 * ["I"] + ["LIF_DI"]]
+ 2 * [3 * ["I"] + ["."]]
+ 4 * [3 * ["F"] + ["LIF_F"]], figsize=(12, 6), dpi=300)
fig.align_ylabels()
plt.subplots_adjust(wspace=0.5, hspace=0.2)
axF_twin = axs["F"].twinx()
for ax, comp in zip([axs["D"], axs["I"], axF_twin], ["D", "I", "F"]):
ax.plot(ref_sed.depth, ref_sed[comp], c="C2", label="ref. process")
ax.set_xticklabels([]) if comp != "F" else None
plot_DIF(sed, axs["D"], axs["I"], axs["F"], time_or_depth="depth", xerr=True, yerr=True, legend=False)
axs["D"].legend(bbox_to_anchor=(0, 0.98), loc="upper left", ncol=3)
plot_DIF(prep_dfs[name + "_4p"], axs["D"], axs["I"], axF_twin, time_or_depth="depth", xerr=True, yerr=True, legend=False, distinguish_subs=False)
plot_DIF(prep_dfs[name + "_2p"], axs["D"], axs["I"], axF_twin, time_or_depth="depth", xerr=True, yerr=True, legend=False, distinguish_subs=False, color="C4")
axF_twin.set_ylabel("Intensity [uT]")
for ax, bs, bs_4p, bs_2p in zip([axs["LIF_DI"], axs["LIF_F"]], [bs_DIs, bs_Fs], [est_bs_DI_4p, est_bs_F_4p], [est_bs_DI_2p, est_bs_F_2p]):
ax.plot(lif_knots, lif(bs[file])(lif_knots), c="C2", lw=2, zorder=2, label="real")
ax.plot(lif_knots, lif(bs_4p)(lif_knots), c="C0", lw=2, zorder=3, label="4p")
ax.plot(lif_knots, lif(bs_2p)(lif_knots), c="C4", lw=2, zorder=3, label="2p")
ax.set_yticks([], [])
axs["LIF_DI"].legend(bbox_to_anchor=(0.5, 1.35), title="Lock-in functions", loc="center", ncol=3)
offsets_text = r"$\bf{Declination\ offsets}$" + "\n"
offsets_text += f"real: A1: {offsets['A1']:.2f}; A2: {offsets['A2']:.2f}\n"
offsets_text += f"4p: A1: {est_offsets_4p['A1']:.2f}; A2: {est_offsets_4p['A2']:.2f}\n"
offsets_text += f"2p: A1: {est_offsets_2p['A1']:.2f}; A2: {est_offsets_2p['A2']:.2f}"
f_shallow_text = r"$\bf{Shallowing\ factor}$" + "\n"
f_shallow_text += f"real: {f_shallow:.3f}\n"
f_shallow_text += f" 4p: {est_f_shallow_4p:.3f}\n"
f_shallow_text += f" 2p: {est_f_shallow_2p:.3f}"
cal_fac_text = r"$\bf{Calibration\ factor}$" + "\n"
cal_fac_text += f"real: {cal_fac:.2f}\n"
cal_fac_text += f" 4p: {est_cal_fac_4p:.2f}\n"
cal_fac_text += f" 2p: {est_cal_fac_2p:.2f}"
axs["D"].text(-0.25, 0.5, offsets_text, transform=axs["D"].transAxes, va="center", ha="center", bbox=props)
axs["I"].text(-0.25, 0.5, f_shallow_text, transform=axs["I"].transAxes, va="center", ha="center", bbox=props)
axs["F"].text(-0.25, 0.5, cal_fac_text, transform=axs["F"].transAxes, va="center", ha="center", bbox=props)
# fig.suptitle(f"Results for {name}", x=0, fontweight="bold")
# plt.savefig(
# f"../../../paper_3/65c4d0ebd16cb35403bef57e/{name}.png",
# bbox_inches="tight",
# )